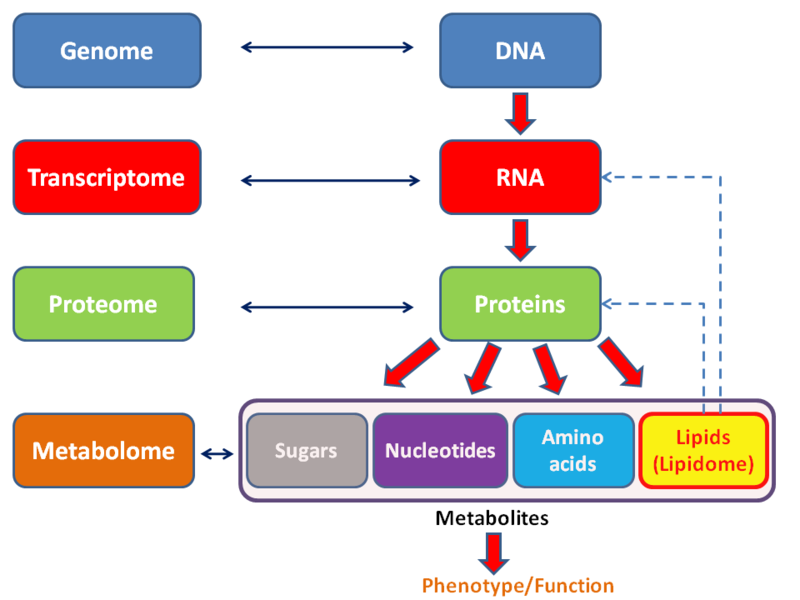
Study of the metabolome.
Some notable examples:
For a commented initial example, see: e. Coli K-12 MG1655 gene thrA.
But BioCyc is generally better otherwise.
I don't think it has any advantage over KEGG however, besides historical interest? Maybe slightly more manual layout and so more beautiful?
Design software for synthetic biological circuit.
The input is in Verilog! Overkill?
Articles by others on the same topic
There are currently no matching articles.