What poor countries have to do to get richer Allow your citizens to have double citizenship by  Ciro Santilli 40 Updated 2025-07-16
Ciro Santilli 40 Updated 2025-07-16
In 2020 Brazil for example, you are not allowed in theory to obtain a double nationality which you were not allowed to have as a birth right.
This means that Brazilian students e.g. in France, many of whom could easily obtain the French citizenship had to either chose between:Can you guess which option Brazilian students would usually pick?
- giving up their Brazilian citizenship. Who the fuck would do that? Brazilians love their country despite all!
- not getting French citizenship. This meant in France having to come 6 AM once a year to some police station on some suburbia to stamp a piece of paper, plus having your VISA completely dependant on your employer for several years until you could obtain a permanent VISA, making it very hard to change jobs, and putting you in a constantly precarious position
- keeping both citizenships, ignoring Brazilian law, which is extremely unlikely to sue you anyways for this bullshit law, and just hope for the best
As a poor country, you have to allow people to obtain multiple citizenship. Students are not going to go back because they don't have the foreign citizenship. They are just going to have to ensure shittier jobs for a few years, thus diminishing the chances that they will actually lean anything useful to bring back to your country later on.
Reviews mostly old RPG and strategy games. And hentai games when it is possible to hide the porn from YouTube. He also sponsors Hentai and uploads it on his website: www.ssethtzeentach.com/nsfw
Ciro Santilli really likes his sense of humor, always going into "politically incorrect" areas, and often making fun of both dictatorships and the USA. He actually knows a bit about politics. Due to the nature of his humour, many of his earlier videos have been taken down from YouTube apparently, www.ssethtzeentach.com/videos mentions:
The videos are also incredibly packed full of well selected edited-in "memes jokes". He particularly likes very short snippets of gay porn which cannot actually be taken down as porn, even though the obviously are porn excerpts, like the buffed dude blowing a kiss to his tit.
www.youtube.com/watch?v=sw8v5__Ytf4 Heroes of Might and Magic III (2018) is one of the best reviews. Sseth likes to find and make fun of game breaking imbalances, something that Ciro likes due to his Ciro Santilli's self perceived creative personality.
www.youtube.com/watch?v=rOJdDvf-tUs The Sseth Streaming Experience (2019), being a live stream, likely shows more realistically how Seth actually talks in real life.
knowyourmeme.com/memes/people/ssethtzeentach gives his profile image from: www.aljazeera.com/features/2012/7/30/the-two-most-loyal-soldiers-in-the-dr-congo
youtu.be/se6Y2o3OqJQ?t=2 shows what could actually be outside of his real window.
Although Ciro Santilli is a big fan of plaintext files and of Vim, not so for games. Games must be easy to understand since they are just a toy.
Tilesets to the rescue!
E.g. in , the underlying field is , the real numbers. And in the underlying field is , the complex numbers.
Any field can be used, including finite field. But the underlying thing has to be a field, because the definitions of a vector need all field properties to hold to make sense.
This shows that viewing electromagnetism as gauge theory does have experimentally observable consequences. TODO understand what that means.
In more understandable terms, it shows that the magnetic vector potential matters where the magnetic field is 0.
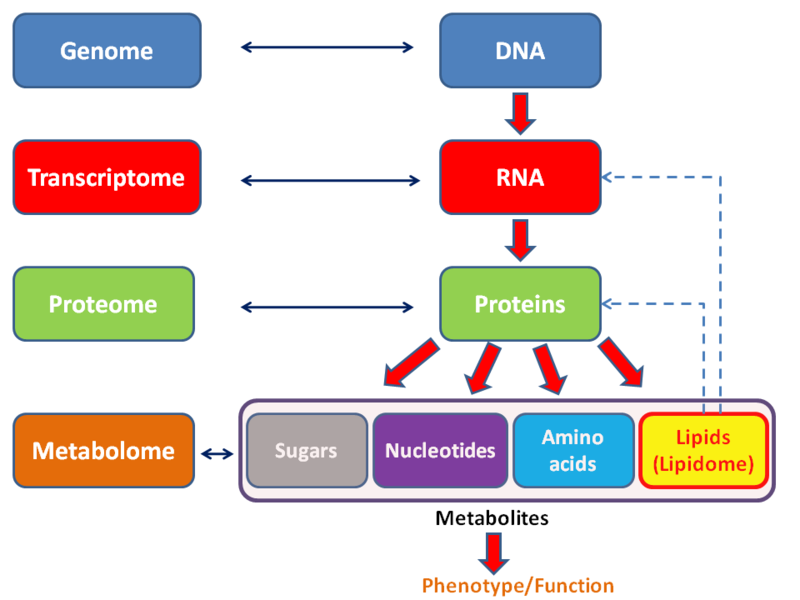
Each of the omics studies a subset of molecular biology with a data intensive and broad point of view that tries to understand global function or organisms, trying to understand what every biologically relevant molecule does as part of the hole metabolism.
The main omics are:
Omics might be stamp collecting, but maybe it is a bit more like Trading card game/Magic: The Gathering collecting, in which the cards that you are collecting actually have specific uses and interactions, especially considering that most metabolic pathways are analogous across many species.
Hierarchical diagram of the major omics
. I guess ended up doing all the "how things should look like" features because they clarify what the website is supposed to do, and I already have my own content to bring it alive via
ourbigbook --web upload.But now I honestly feel that all the major elements of "how things should look like" have fallen into place.
I was really impressed by Trillium Notes. I should have checked it long ago. The UI is amazing, and being all Js-based, could potentially be reused for our purposes. The project itself is a single-person/full trust notetaking only for now however, so not a direct replacement to OurBigBook.
- upload all of cirosantilli.com to ourbigbook.com. I will do this by implementing an import from filesystem functionality based on the OurBigBook CLI. This will also require implementing slit headeres on the server to work well, I'll need to create one
Articlefor every header on render. - get
\xand\Includeworking on the live web preview editor. This will require creating a new simple API, currently the editor jus shows broken references, but final render works because it goes through the database backend - implement email verification signup. Finally! Maybe add some notifications too, e.g. on new comments or likes.
A non-tool-assisted speedrun.
Ciro Santilli views humans as biological robots, and therefore RTA videos can be thought of as probabilistic TAS with human achievable reflex constraints.
This aspect is especially highlighted in "speed run record evolution videos", which can be quite fun, e.g. www.youtube.com/watch?v=pmS9e7kzgS4 Ocarina of Time - World Record History and Progression (Any% Speedrun, 1990s-2017) by retro (2017)
From a similar point of view, Ciro also sometimes watches/learns a bit about competitive PvP games from a "could a computer play this better than a human" point of view.
Ciro also likes to watch commented manual speedruns of games as a way of experiencing the game at a high level without spending too much time on it, often from Games Done Quick. Their format is good because it generally showcases one player focusing more on the gameplay, and three couch commentators to give context, that's a good setup.
It is a
Unlisted articles are being shown, click here to show only listed articles.