For the strong.
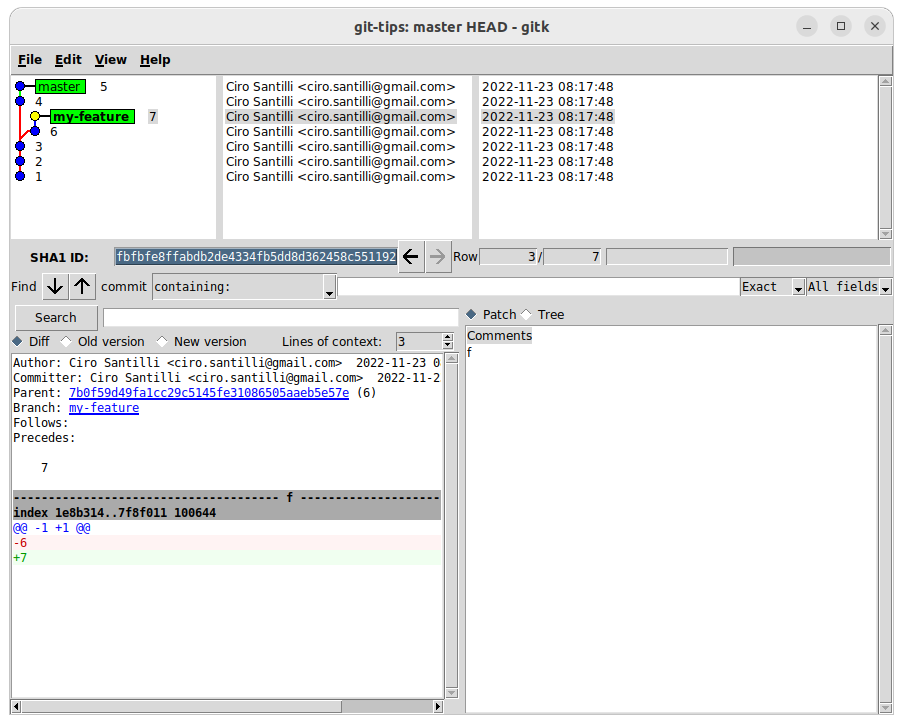
git log --abbrev-commit --decorate --graph --pretty=oneline master HEADOutput:
* b4ec057 (master) 5
* 0b37c1b 4
| * fbfbfe8 (HEAD -> my-feature) 7
| * 7b0f59d 6
|/
* 661cfab 3
* 6d748a9 2
* c5f8a2c 1If we also add the As we can see, this removes any commit that is neither:
--simplify-by-decoration, which you very often want want on a real repository with many commits:* b4ec057 (master) 5
| * fbfbfe8 (HEAD -> my-feature) 7
|/
* c5f8a2c 1- under a branch or tag
- at the intersection of too branches or tags
In order to solve conflicts, you just have to understand what commit you are trying to move where.
E.g. if from:we do:what happens step by step is first 6 is moved on top of 5:and then 7 is moved on top of the new 6:
5 master
|
4 7 my-feature HEAD
| |
3 6
|/
2
|
1git rebase master6on5 HEAD
|
5 master
|
4 7 my-feature
| |
3 6
| |
2-----------------+
|
17on5 HEAD
|
6on5
|
5 master
|
4 7 my-feature
| |
3 6
| |
2-----------------+
|
17on5 my-feature HEAD
|
6on5
|
5 master
|
4
|
3
|
2
|
1 Git tips The key to solve conflicts: see the two conflicting diffs by  Ciro Santilli 40 Updated 2025-07-16
Ciro Santilli 40 Updated 2025-07-16
The key to solve conflicts is:
You have to understand what are the two commits that touched a given line (one from master, one from features), and then combine them somehow.
CLI hello world:
gnuplot -p -e 'p sin(x)'External 3D view of the Brodmann areas
. Source. The bad:
- Clunky UI
- circuit diagram doesn't show any state??
Likely the best JavaScript 2D game engine as of 2023.Uses Matter.js as a physics engine if enabled. There's also an alternative (in-house?) "arcade" engine: photonstorm.github.io/phaser3-docs/Phaser.Physics.Arcade.ArcadePhysics.html but it appears to be simpler/less robust (but also possibly faster).
- github.com/photonstorm/phaser
- phaser.io/
- phaser.io/examples/v3.85.0/games contains the demo games
There are unlisted articles, also show them or only show them.