Brain in a vat Updated 2025-07-16
The ideal university Updated 2025-07-16
This is Ciro Santilli's ideal university system. It is a system that actually lives up to the name "Open University":
- no enrolment, no prerequisites. Exam as a service examination style, likely free to anyone who wants to take them, only to determine:
- who gets to use physical facilities, notably laboratories
- which students do you want to pick as apprentices/workers/PhDs
- no tuition fees: free gifted education
- school must offer free accommodation for students
- force teachers to publish their teaching material with an open license
- how to teach
Related:
- reallyopenuniversity.wordpress.com/what-is-the-rou/
Apparently Leeds based. Focused only on student fees seemingly, not how to solve it with tech/efficiency:
So far as of the early 2020's, the university that comes closest to some but not all of these principles is the University of the People. It's sad that it's such a crappy unknown thing, but it is what it is.
Yitang Zhang's theorem Updated 2025-07-16
There are infinitely many primes with a neighbor not further apart than 70 million. This was the first such finite bound to be proven, and therefore a major breakthrough.
This implies that for at least one value (or more) below 70 million there are infinitely many repetitions, but we don't know which e.g. we could have infinitely many:or infinitely many:or infinitely many:or infinitely many:but we don't know which of those.
The Prime k-tuple conjecture conjectures that it is all of them.
Braindumping Updated 2025-07-16
There are two ways:
- manually dumping your brain on media such as notebooks, wikis or videos, i.e. forming a personal knowledge base
- technically using sensors for brain scanning
CIA 2010 covert communication websites Expired domain trackers Updated 2025-07-16
When you Google most of the hit domains, many of them show up on "expired domain trackers", and above all Chinese expired domain trackers for some reason, notably e.g.:This suggests that scraping these lists might be a good starting point to obtaining "all expired domains ever".
- hupo.com: e.g. static.hupo.com/expdomain_myadmin/2012-03-06(国际域名).txt. Heavily IP throttled. Tor hindered more than helped.Scraping script: ../cia-2010-covert-communication-websites/hupo.sh. Scraping does about 1 day every 5 minutes relatively reliably, so about 36 hours / year. Not bad.Results are stored under
tmp/humo/<day>.Check for hit overlap:The hits are very well distributed amongst days and months, at least they did a good job hiding these potential timing fingerprints. This feels very deliberately designed.grep -Fx -f <( jq -r '.[].host' ../media/cia-2010-covert-communication-websites/hits.json ) cia-2010-covert-communication-websites/tmp/hupo/*There are lots of hits. The data set is very inclusive. Also we understand that it must have been obtains through means other than Web crawling, since it contains so many of the hits.Some of their files are simply missing however unfortunately, e.g. neither of the following exist:webmasterhome.cn did contain that one however: domain.webmasterhome.cn/com/2012-07-01.asp. Hmm. we might have better luck over there then?2018-11-19 is corrupt in a new and wonderful way, with a bunch of trailing zeros:ends in:wget -O hupo-2018-11-19 'http://static.hupo.com/expdomain_myadmin/2018-11-19%EF%BC%88%E5%9B%BD%E9%99%85%E5%9F%9F%E5%90%8D%EF%BC%89.txt hd hupo-2018-11-19000ffff0 74 75 64 69 65 73 2e 63 6f 6d 0d 0a 70 31 63 6f |tudies.com..p1co| 00100000 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00 |................| * 0018a5e0 00 00 00 00 00 00 00 00 00 |.........|More generally, several files contain invalid domain names with non-ASCII characters, e.g. 2013-01-02 contains365<D3>л<FA><C2><CC>.com. Domain names can only contain ASCII charters: stackoverflow.com/questions/1133424/what-are-the-valid-characters-that-can-show-up-in-a-url-host Maybe we should get rid of any such lines as noise.Some files around 2011-09-06 start with an empty line. 2014-01-15 starts with about twenty empty lines. Oh and that last one also has some trash bytes the end<B7><B5><BB><D8>. Beauty. - webmasterhome.cn: e.g. domain.webmasterhome.cn/com/2012-03-06.asp. Appears to contain the exact same data as "static.hupo.com"Also has some randomly missing dates like hupo.com, though different missing ones from hupo, so they complement each other nicely.Some of the URLs are broken and don't inform that with HTTP status code, they just replace the results with some Chinese text 无法找到该页 (The requested page could not be found):Several URLs just return length 0 content, e.g.:It is not fully clear if this is a throttling mechanism, or if the data is just missing entirely.
curl -vvv http://domain.webmasterhome.cn/com/2015-10-31.asp * Trying 125.90.93.11:80... * Connected to domain.webmasterhome.cn (125.90.93.11) port 80 (#0) > GET /com/2015-10-31.asp HTTP/1.1 > Host: domain.webmasterhome.cn > User-Agent: curl/7.88.1 > Accept: */* > < HTTP/1.1 200 OK < Date: Sat, 21 Oct 2023 15:12:23 GMT < Server: Microsoft-IIS/6.0 < X-Powered-By: ASP.NET < Content-Length: 0 < Content-Type: text/html < Set-Cookie: ASPSESSIONIDCSTTTBAD=BGGPAONBOFKMMFIPMOGGHLMJ; path=/ < Cache-control: private < * Connection #0 to host domain.webmasterhome.cn left intactStarting around 2018, the IP limiting became very intense, 30 mins / 1 hour per URL, so we just gave up. Therefore, data from 2018 onwards does not contain webmasterhome.cn data.Starting from2013-05-10the format changes randomly. This also shows us that they just have all the HTML pages as static files on their server. E.g. with:we see:grep -a '<pre' * | s2013-05-09:<pre style='font-family:Verdana, Arial, Helvetica, sans-serif; '><strong>2013<C4><EA>05<D4><C2>09<C8>յ<BD><C6>ڹ<FA><BC><CA><D3><F2><C3><FB></strong><br>0-3y.com 2013-05-10:<pre><strong>2013<C4><EA>05<D4><C2>10<C8>յ<BD><C6>ڹ<FA><BC><CA><D3><F2><C3><FB></strong> - justdropped.com: e.g. www.justdropped.com/drops/010112com.html. First known working day:
2006-01-01. Unthrottled. - yoid.com: e.g.: yoid.com/bydate.php?d=2016-06-03&a=a. First known workding day:
2016-06-01.
Data comparison:
- 2012-01-01Looking only at the
.com:The lists are quite similar however.- webmastercn has just about ten extra ones than justdropped, the rest is exactly the same
- justdropped has some extra and some missing from hupo
We've made the following pipelines for hupo.com + webmasterhome.cn merging:
./hupo.sh &
./webmastercn.sh &
./justdropped.sh &
wait
./justdropped-post.sh
./hupo-merge.sh
# Export as small Google indexable files in a Git repository.
./hupo-repo.sh
# Export as per year zips for Internet Archive.
./hupo-zip.sh
# Obtain count statistics:
./hupo-wc.shCount unique domains in the repos:
( echo */*/*/* | xargs cat ) | sort -u | wcThe extracted data is present at:Soon after uploading, these repos started getting some interesting traffic, presumably started by security trackers going "bling bling" on certain malicious domain names in their databases:
- archive.org/details/expired-domain-names-by-day
- github.com/cirosantilli/expired-domain-names-by-day-* repos:
- github.com/cirosantilli/expired-domain-names-by-day-2006
- github.com/cirosantilli/expired-domain-names-by-day-2007
- github.com/cirosantilli/expired-domain-names-by-day-2008
- github.com/cirosantilli/expired-domain-names-by-day-2009
- github.com/cirosantilli/expired-domain-names-by-day-2010
- github.com/cirosantilli/expired-domain-names-by-day-2011 (~11M)
- github.com/cirosantilli/expired-domain-names-by-day-2012 (~18M)
- github.com/cirosantilli/expired-domain-names-by-day-2013 (~28M)
- github.com/cirosantilli/expired-domain-names-by-day-2014 (~29M)
- github.com/cirosantilli/expired-domain-names-by-day-2015 (~28M)
- github.com/cirosantilli/expired-domain-names-by-day-2016
- github.com/cirosantilli/expired-domain-names-by-day-2017
- github.com/cirosantilli/expired-domain-names-by-day-2018
- github.com/cirosantilli/expired-domain-names-by-day-2019
- github.com/cirosantilli/expired-domain-names-by-day-2020
- github.com/cirosantilli/expired-domain-names-by-day-2021
- github.com/cirosantilli/expired-domain-names-by-day-2022
- github.com/cirosantilli/expired-domain-names-by-day-2023
- github.com/cirosantilli/expired-domain-names-by-day-2024
- GitHub trackers:
- admin-monitor.shiyue.com
- anquan.didichuxing.com
- app.cloudsek.com
- app.flare.io
- app.rainforest.tech
- app.shadowmap.com
- bo.serenety.xmco.fr 8 1
- bts.linecorp.com
- burn2give.vercel.app
- cbs.ctm360.com 17 2
- code6.d1m.cn
- code6-ops.juzifenqi.com
- codefend.devops.cndatacom.com
- dlp-code.airudder.com
- easm.atrust.sangfor.com
- ec2-34-248-93-242.eu-west-1.compute.amazonaws.com
- ecall.beygoo.me 2 1
- eos.vip.vip.com 1 1
- foradar.baimaohui.net 2 1
- fty.beygoo.me
- hive.telefonica.com.br 2 1
- hulrud.tistory.com
- kartos.enthec.com
- soc.futuoa.com
- lullar-com-3.appspot.com
- penetration.houtai.io 2 1
- platform.sec.corp.qihoo.net
- plus.k8s.onemt.co 4 1
- pmp.beygoo.me 2 1
- portal.protectorg.com
- qa-boss.amh-group.com
- saicmotor.saas.cubesec.cn
- scan.huoban.com
- sec.welab-inc.com
- security.ctrip.com 10 3
- siem-gs.int.black-unique.com 2 1
- soc-github.daojia-inc.com
- spigotmc.org 2 1
- tcallzgroup.blueliv.com
- tcthreatcompass05.blueliv.com 4 1
- tix.testsite.woa.com 2 1
- toucan.belcy.com 1 1
- turbo.gwmdevops.com 18 2
- urlscan.watcherlab.com
- zelenka.guru. Looks like a Russian hacker forum.
- LinkedIn profile views:
- "Information Security Specialist at Forcepoint"
Check for overlap of the merge:
grep -Fx -f <( jq -r '.[].host' ../media/cia-2010-covert-communication-websites/hits.json ) cia-2010-covert-communication-websites/tmp/merge/*Next, we can start searching by keyword with Wayback Machine CDX scanning with Tor parallelization with out helper ../cia-2010-covert-communication-websites/hupo-cdx-tor.sh, e.g. to check domains that contain the term "news":produces per-year results for the regex term OK lets:
./hupo-cdx-tor.sh mydir 'news|global' 2011 2019news|global between the years under:tmp/hupo-cdx-tor/mydir/2011
tmp/hupo-cdx-tor/mydir/2012./hupo-cdx-tor.sh out 'news|headline|internationali|mondo|mundo|mondi|iran|today'Other searches that are not dense enough for our patience:
world|global|[^.]infoOMG and a few more. It's amazing.
news search might be producing some golden, golden new hits!!! Going full into this. Hits:- thepyramidnews.com
- echessnews.com
- tickettonews.com
- airuafricanews.com
- vuvuzelanews.com
- dayenews.com
- newsupdatesite.com
- arabicnewsonline.com
- arabicnewsunfiltered.com
- newsandsportscentral.com
- networkofnews.com
- trekkingtoday.com
- financial-crisis-news.com
Ciro Santilli's children Updated 2025-07-16
Count: 0.
Lumai Updated 2025-07-16
Funding:
- 2023: 1.1m pounds www.uktech.news/deep-tech/lumai-grant-20230215
The IT Crowd Updated 2025-07-19
The Legend of Zelda: Ocarina of Time Updated 2025-07-16
This game was mind blowing to Ciro Santilli and all kids. It felt so real. The perfect contrast between peaceful town work and saving the world. OMG.
Breakthrough Prize Updated 2025-07-16
By Zuckerberg. The selection seems decent. And natural sciences only, which is good. A bit more application oriented than the Nobel Prize it seems, e.g. 2022 separates physics and fundamental physics.
Appears to explain award reasoning even worse than the Nobel Foundation.
Brian Josephson Updated 2025-07-16
The World Of Enrico Fermi by Harvard Project Physics (1970) Updated 2025-07-16
torchvision ResNet Updated 2025-07-16
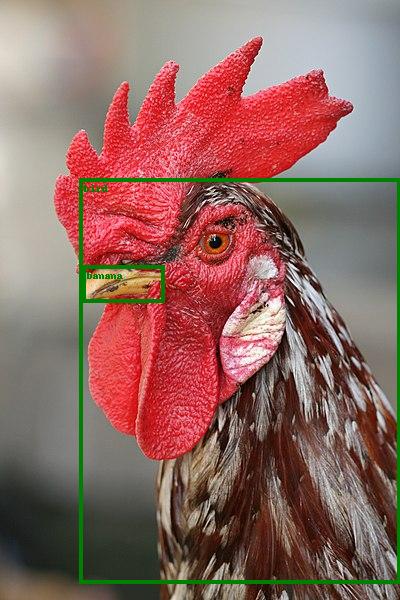
pytorch.org/vision/0.13/models.html has a minimal runnable example adapted to python/pytorch/resnet_demo.py.
That example uses a ResNet pre-trained on the COCO dataset to do some inference, tested on Ubuntu 22.10:This first downloads the model, which is currently 167 MB.
cd python/pytorch
wget -O resnet_demo_in.jpg https://upload.wikimedia.org/wikipedia/commons/thumb/6/60/Rooster_portrait2.jpg/400px-Rooster_portrait2.jpg
./resnet_demo.py resnet_demo_in.jpg resnet_demo_out.jpgWe know it is COCO because of the docs: pytorch.org/vision/0.13/models/generated/torchvision.models.detection.fasterrcnn_resnet50_fpn_v2.html which explains that is an alias for:
FasterRCNN_ResNet50_FPN_V2_Weights.DEFAULTFasterRCNN_ResNet50_FPN_V2_Weights.COCO_V1After it finishes, the program prints the recognized classes:so we get the expected
['bird', 'banana']bird, but also the more intriguing banana. Brightest natural objects in the sky Updated 2025-07-16
British Overseas Territories Updated 2025-07-16
British physics research institute Updated 2025-07-16
Tinker Tailor Soldier Spy (film) Updated 2025-07-16
This is not bad, but some divergences to the better BBC miniseries, which presumably sticks more closely to the novel:
- in the film Jim Prideaux is captured in a cafe in Prague, in the series it's in the woods. It is therefore much more plausible that he would have been shot.
- in the film Peter Guillam is played by Benedict Cumberbatch, who feels a bit young to be Ricki Tarr's boss. Not impossible, but still.
- the series is much less chronological, and more flashback based, as new information becomes available. The film is more chronological, which makes it easier to understand, but less interesting at the same time.
- in the film they shoot the Russian girl Irina in front of Jim, in the series the fact that she was shot is only known through other sources. The film has more eye candy, which weakens it.
- Toby Esterhase is not threatened in an airfield, only in a safe ;house in London.
How large primes are found for RSA Updated 2025-07-16
Answers suggest hat you basically pick a random large odd number, and add 2 to it until your selected primality test passes.
The prime number theorem tells us that the probability that a number between 1 and is a prime number is .
Human Compatible Updated 2025-07-16
The key takeaway is that setting an explicit value function to an AGI entity is a good way to destroy the world due to poor AI alignment. We are more likely to not destroy by creating an AI whose goals is to "do want humans what it to do", but in a way that it does not know before hand what it is that humans want, and it has to learn from them. This approach appears to be known as reward modeling.
Some other cool ideas:
- a big thing that is missing for AGI in the 2010's is some kind of more hierarchical representation of the continuous input data of the world, e.g.:
- game theory can be seen as part of artificial intelligence that deals with scenarios where multiple intelligent agents are involved
- probability plays a crucial role in our everyday living, even though we don't think too much about it every explicitly. He gives a very good example of the cost/risk tradeoffs of planning to the airport to catch a plane. E.g.:
- economy, and notably the study of the utility, is intrinsically linked to AI alignment
Human essential fatty acid Updated 2025-07-16
There are unlisted articles, also show them or only show them.