Why is COVID-19 so serious in some people but not in others? by  Ciro Santilli 40 Updated 2025-07-16
Ciro Santilli 40 Updated 2025-07-16
There are a few possibilities:
- genetics
- bibliography:
- www.science.org/doi/10.1126/scitranslmed.abj7521 Identification of driver genes for critical forms of COVID-19 in a deeply phenotyped young patient cohort by Carapito et al. (2021)
- bibliography:
- state of the immune system based on disease history
- age
- www.youtube.com/watch?v=nrBiDRZRK5g Maxwell Lagrangian Derivation by Dietterich Labs (2019)
- www.youtube.com/watch?v=yo-Z3RO-eeY Deriving the Maxwell Lagrangian by Pretty Much Physics (2019)
Cool data embedded in the Bitcoin blockchain Base58 messages by  Ciro Santilli 40 Updated 2025-07-16
Ciro Santilli 40 Updated 2025-07-16
Bitcoin addresses are by convention expressed in Base58, which is a human readable binary-to-text encoding invented by Bitcoin.
It is a bit like Base64, but obsessed with eliminating characters that look like one another in popular but stupid fonts like capital "I" and lower case ell "l". As such, any embedded text is rather obfuscated due to this limitations, and people often resort to leet-like replacements such as '1' to represent 'I'.
This seems to be one of the earliest strategies used to encode messages into the Bitcoin blockchain. The first known example appears in 2011. Then starting November 2011, a large number of messages were inscribed n short successsion, presumably by a single person or small group.
The interest in Base58 encoding might have initially arisen with people's desire to have "vanity addresses", that is Bitcoin addresses that have real words in them, much like vanity plates or vanity numbers. Such addresses with long words in them are hard to find while keeping the address spendable, because they have to correspond to a private key. An extreme notable example is:which contains the awkward 13 letter word:in it. TODO: proof that it is pendable?
embarrassable
Perhaps inspired by this, some people also decided to use Base58 addresses as a way to create more general unspendable inscriptions, even even though the method is much more clumsy and complicated than P2FKHS. There is however a certain art to working under limitations.
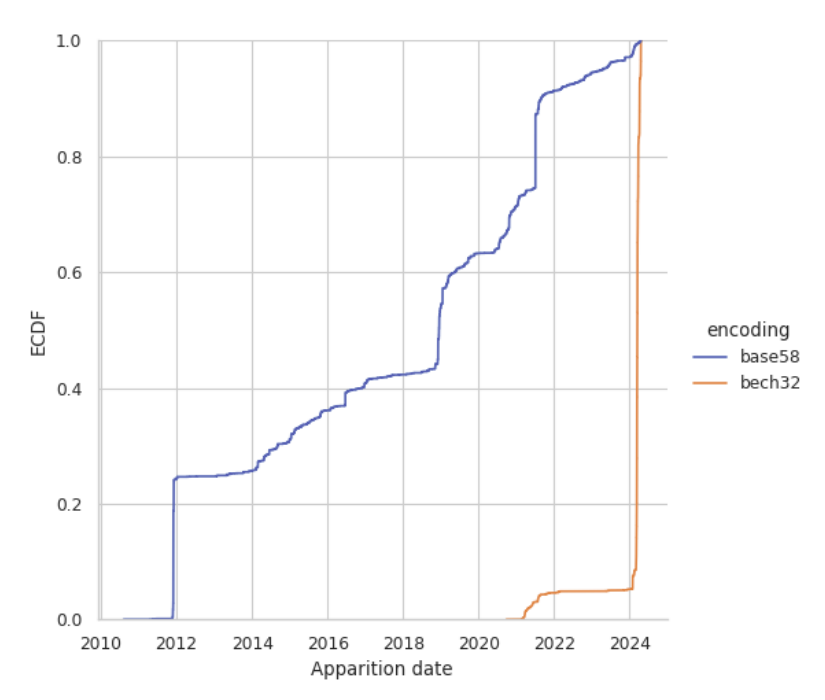
Total burn addresses as a function of time found by Bitcoin Burn Addresses: Unveiling the Permanent Losses and Their Underlying Causes
. Although it is not solely focused on inscriptions and may also contain functional burn addresses, it is likely that the methods of Khatib/Legout capture the overall trend of base58 inscription counts.These messages were originally found with: github.com/cirosantilli/bitcoin-inscription-indexer#payload-size-out-utxo-2vals which tracks the largest transactions with unspent outputs.
Bitcoin Burn Addresses: Unveiling the Permanent Losses and Their Underlying Causes later revealed many new ones.
Bitcoin Burn Addresses: Unveiling the Permanent Losses and Their Underlying Causes later revealed many new ones.
Finding Base58 messages is intrinsically hard for a few reasons
- the words may be garbled by Base58 leet
- only very small ammounts of data can be encoded at a time, and all of it contains ASCII, so you can't just "find all long ASCII strings" as we started doing for other ASCII inscriptions a la
strings -n20; you have to use some dictionary as a basis - the Base58 does not show up raw on the blockchain, as it is just a human representation for the actual binary data that does, so you can't just strings the blockchain, you have to parse it
The interesting following transactions contain base58 encoded messages on addresses, sorted chronologically, and heighlighted either due to their earliness or historical or artistic quality:
- on two transactions of block 124726 (2011-05-18) someone created two addresses whose base58 differs only by one character,
Win one is replaced byxin he other:bitcointalk.org/index.php?topic=20955.msg264038#msg264038 How is this address possible? (2011-06-22) user ByteCoin suggests that this was done to highlight the fact that the checksum at the end of base58 addresses against 1 character changes. They also highlight another pair where addresses are equal except for two adjacent swapped characters:18->81but these are not present in the blockchain itself.1ByteCoinAddressesMatchcNN781jjwLY 1ByteCoinAddressesMatchcNN718jjwLY - Around July 2011 there seems to have been a surge of interest in vanity addresses, and it appears that someone was "squatting" long lists of interesting addresses that they managed to generate for later sale. These addresses are present in the hundreds in a few transactions chains, and they do not seem to contain any coherent messages across the outputs. Most encode given names, which would be the easiest type of address to sell. This theory is proposed e.g. at: bitcointalk.org/index.php?topic=84569.msg992950#msg992950 and it seems as the most plausible one to us. An example of this is tx acdd81bab63ee42e28296dd5c21e8a29392e409026fc206acf5931b12a31141d block 136273 (2011-07-14) which starts off with:For the purposes of this museum, this is a noteworty event, but it has little artistic value for large ammounts of bulk, and therefore also serves as noise that must be removed if we want to find other more personal and varied inscriptions. We will keep a list of such transactions at: Section "Bitcoin 2011 vanity address pool".
1MeNDez2hmZoehh5JAtS2ZJQfAFZFfSQSi 1ALonzoPwyf8CNVQnVNXNBjacPXaUdZGgm 1MattieiicNRfse5jTVU2X8pX6Cyr7BZVR 1TraciFRboW661p1LfRaULwwefeo8KtQa 1MartinHaferkorncii112o11HdkMrtttDon tx dab55eefd5cef495719a43bbd190c57c8ca60ecc45d630edf3442b2096965a97 block 152851 (2011-11-11) encodes the name:Likely:Martin Haferkorn
1EricLombrozoXXXXXXXXXXXXXXXWACBVBappears on 3 separate transactions on 2011-11-24:Alsmost certainly this guy:www.officialusa.com/names/Sandra-Sandic/ suggests a link between Eric and Sandra sharing phone number (858) 461-1843 and residing at 12631 El Camino Real, San Diego, CA. Eric's LinkedIn marks him as living in San Diego, and Sandra's birthday is marked 1969-01-05, so matching the inscription year. The address shows as a regular appartment block on Google Maps, so maybe they are not crazy rich, or they have restraint. besthistorysites.net/name/eric-lombrozo reconfirms the address.- block 154630:
- block 154637: tx dea183908e40e0cebfee6a0d8362b299e07cf193fbc02ffd3308b43781eca208. This one is more interesting and also contains a second output, both at 0.005 BTCso possibly a wedding token of Eric with Sandra Sandic after two previous test transactions. This also possibly gives Sandra's birth year of 1969. Pinged him at: x.com/cirosantilli/status/1904212575211901129.
1969SandraSandicXXXXXXXXXXXXXvdEiU
In 2023 this Sandra Sandic on Facebook liked this post related to a show in San Diego, giving a possible profile. At this post she links to this story about Erik Finman, young Bitcoin millionaire, thus establishing an interest link between that profile and Bitcoin. She also has various posts in Bosnian, so she speaks the language and is likely a first generation immigrant.1BitTaLkTVChristmasSpeciaLXXRix9Eais repeated a dozen times on transactions between tx 8e2bacf9971ce1a29d69d1a0484bfaa198257cc116530c7415ab6c38ae54ebc3 block 154721 (2011-11-25) and 2011-11-27.It is a quick ad for the BitTalk.tv Christmas Special by Matthew N. Wright. bitcointalk.org/index.php?topic=3025298.0 mentions he is the founder bittalk.tv and co-founder of Bitcoin Magazine. TODO is the video still watchable somewhere? Also announced at: bitcointalk.org/index.php?topic=52712. As of 2025, the domain had been reappropriated as a SolarMovie mirror. It is quite likely that all the large set of message that follow were inscribed by him. Related:- www.reddit.com/r/Bitcoin/comments/ruo73/matthew_n_wright_scammer_of_bitcoinmagazine_and/ Matthew N. Wright, scammer of BitcoinMagazine and BBBB (2012)
- www.reddit.com/r/Bitcoin/comments/znhsj/matthew_n_wright_apologizes/ Matthew N. Wright "apologizes" (2012)
Later on there is also another variant addresses11Bitta1ktvchristmasspecia1WNDvAaon repeated almost 300 times on tx ace9524519577138ca98ec01651758fd1e5ec33ce0110c6681eccba0e716cc7a block 155545 (2011-12-01)Other likely mentions of Matthew N Wright:11MatthewLovesmandaXXXXXXXXabCJPYon tx d7c57205d69420dc7f4593b4de0806c9ec96f4755b64315cd034bd4b0b90dc2a block 155698 (2011-12-01) has a quick love declaration:Matthew loves Amanda
1MatthewNWrightisaScammer124DNsfXis etched twice
- tx 28ccf29cfcc9f82d42793db770e7c7894d61ccf3d18299f34bda2e54415da287 block 154769 (2011-11-25) contains a short excerpt from Alice in WonderlandOriginal text referred to:
1But1DontWantToGoAmongMadxxxzDmyW6 1Peop1eA1iceRemarkedxxxxxxxxxuLyKu 12ohYouCantHe1pThatxxxxxxxxxzCjyMs 19SaidTheCatWereA11MadHerexxyTvEir 191mMadYoureMadxxxxxxxxxxxxxvwA4Up 1HowDoYouKnow1mMadSaidA1icexxZA4Nr 12YouMustBeSaidTheCatxxxxxxxz2tFa2 12orYouWou1dntHaveComeHerexxvtHbqq - tx 3bbd94d22346a3bfb44257293e10c3b5c9ee39230c1cd358bdce2bf03c61ba0b (block 154965 , 2011-11-27) contains 49 base58 messages on a single transaction transcribing this version of the Emerald Tablet, a type of mystical medieval text:Some of the messages weirdly have "xoxo" inserted into them, not sure why, e.g.
12TisTrueWithoutALie22222221wT3qjn 1CertainAndMostTrue2222222225YPnJF 12ThatWhich1sBe1ow1sAs222221y3G7mv 12ThatWhich1sAboveAnd2222221vxkcEq ... 1AboutTheWorkingsofTheSun1zzyWJtfmThe full decoded text is:12TheFatherofTheWho1eWor1d2249xs5g 191sHereXoXoXoXoXoXoXoXoXoXo72uqJv 191tsPower1sWho1e1f1tHasXoXoWhYr3M 1BeenTurned1ntoEarth222222221soWALIt is true, without error, certain and most true,
That which is below is as that which is above, and that which is above is as that which is below, to perform the miracles of the one thing.
And as all things were from the one, by means of the meditation of the one, thus all things were born from the one, by means of adaptation.
Its father is the Sun, its mother is the Moon, the Wind carried it in its belly, its nurse is the earth.
The father of the whole world is here.
Its power is whole if it has been turned into earth.
You will separate the earth from the fire, the subtle from the dense, sweetly, with great skill.
It ascends from earth into heaven and again it descends to the earth, and receives the power of higher and of lower things.
Thus you will have the Glory of the whole world.
Therefore will all obscurity flee from you.
Of all strength this is true strength, because it will conquer all that is subtle, and penetrate all that is solid.
Thus was the world created.
From this were wonderful adaptations, of which this is the means. Therefore am I named Thrice-Great Hermes, having the three parts of the philosophy of the whole world.
It is finished, what I have said about the working of the Sun. 11111111LeonhardEu1er111126nxjPon tx 80ddf2e7e04922e2cbf6e744dbf47aec02d781505d8b2c4ee5f725b8882ddb2d block 155051 (2011-11-28) is a tribute to Swiss mathematician Leonhard EulerFigure 3. Swiss mathematician Leonard Euler. Source. Off-chain image.- 024b093afb54f69426c5624f09a5f2d3791ce20513225cbb42d333ad72f8576e block 155256 (2011-11-29) has two self-explanatory outputs:
112JustATwoLine222222222221vcJxpZ 11111Test111112222222222222LiApa - tx 8f64d2b7a762767e3870c4aee95f8c7b5439cf02cf7d7e5d99b6e39967ecada8 block 155256 (2011-11-29) encodes the poem "Shall I compare thee to a summer's day?" by Shakespeare 22 addresses starting with:Full original text:
11Sha111CompareTheeToAXXXXXVnRohE 11SummersDayThouArtMoreXXXXUcpgnX 11Love1yAndMoreTemperateXXXUu485j ...More Shakespeare follows at tx 0ae2eaaa9cddafba89b4c92d074f4e5254cbf7691cbe7f64660bf549c7071147 block 155383 (2011-11-20) has a passage from Romeo and Juliet starting with:Shall I compare thee to a summer's day?
Thou art more lovely and more temperate.
Rough winds do shake the darling buds of May,
And summer's lease hath all too short a date.
Sometime too hot the eye of heaven shines,
And often is his gold complexion dimmed;
And every fair from fair sometime declines,
By chance, or nature's changing course, untrimmed;
But thy eternal summer shall not fade,
Nor lose possession of that fair thou ow'st,
Nor shall death brag thou wand'rest in his shade,
When in eternal lines to Time thou grow'st.
So long as men can breathe, or eyes can see,
So long lives this, and this gives life to thee.Full original text:11TisButThyNameThat1sMyXXXXWabTZh 11EnemyThouArtThyse1fThoughXNRG4J 11NotAMontagueWhatsMontagueYEJDfM 111t1sNorHandNorFootNorArmXVNeFEV'Tis but thy name that is my enemy;
Thou art thyself, though not a Montague.
What's Montague? It is nor hand, nor foot,
Nor arm, nor face, nor any other part
Belonging to a man. O, be some other name!
What's in a name? That which we call a rose,
By any other word would smell as sweet.
So Romeo would - were he not Romeo called -
Retain that dear perfection which he owes
Without that title. Romeo, doff thy name,
And for that name, which is no part of thee,
Take all myself. - Various other notable texts follow on 2011-12-01:
- tx 1f9606f267cc398356663b14d1a7a3591e3da06572893394c14975a6fc11798f block 155467 (2011-12-01) contains an excerpt from Newton's Principia starting with:Full original text[ref]:
11Ru1e1WeAreToAdmitNoMoreXXazQ96z 11CausesofNatura1ThingsThanZAQ9ig 11SuchAsAreBothTrueAndXXXXXZyzfQp 11SufficientToExp1ainTheirXVSC2gYRULE 1 We are to admit no more causes of natural things, than such as are both true and sufficient to explain their appearances.RULE III The qualities of bodies, which admit neither intension nor remission of degrees, and which are found to belong to all bodies within reach of our experiments, are to be esteemed the universal qualities of all bodies whatsoever.RULE IV In experimental philosophy we are to look upon propositions collected by general induction from phenomena as accurately or very nearly true, notwithstanding any contrary hypotheses that may be imagined, till such time as other phenomena occur, by which they may either be made more accurate, or liable to exceptions. - tx 89010c791c9d7ed24affa1d638b12179d2ca7ec91704fe906834386f43a8101d starting at
11When1nTheCourseofHumanXXXXdfMdQ: Declaration of Independence - tx f7ca83a8a2e1c78efdfde0791d99a567ddaa60805c3b5b857bc7ec14ec2c8204 starting at
11AVa1id1dentifier1sAXXXXXXcrnyki: likely contains an excerpt of the C or C++ standard. Possible source: en.wikipedia.org/wiki/C_data_types. - tx 028b8514a4f6cc96ac3c1c83dbb117ab9dc5eb09deab7b49bf038fd460173127 starting at
11TheSetupTheJokeA1waysXXXXTF9Wzp: The aristocrats by HP Lovecraft, which talks about the The Aristocrats joke pattern - tx bd513d9ee605ead1a299c9dfb77de1127bf651c54d99820e9be8b40cef8c8dfe starting at
11Si1ex1sAnAcronymForXXXXXXcujTa5talks briefly about SILEX (separation of isotopes by laser excitation) - tx ef374dcc5b23f16ecb0b1b639ba577d2acda7ad32321b5866db2fa9e6807b9c5 block 155494 (2011-12-01) contains the intruction from the bitcoin.org website: web.archive.org/web/20210129054851/https://bitcoin.org/en/
11Bitcoin1sADecentra1izedXXWPM6Hs 11PeertopeerNetworkoverXXXXUkyy3M 11WhichUsersMakeXXXXXXXXXXXX4tQgN 11TransactionsThatAreXXXXXXVdZfnJ - tx 05ee60dfb92795c79e46e106f52bbdbc1006eba0837ed9e4ad99d9b214eb5fcf block 155538 contains a tribute to Archimedes:original text at: mathshistory.st-andrews.ac.uk/Biographies/Archimedes/
11ArchimedesWasANativeofXXXXJsj6W 11SyracuseSici1y1t1sXXXXXXXaYF4CE 11ReportedBySomeAuthorsThatXjFBcV 11HeVisitedEgyptAndThereXXXYVzj58Archimedes was a native of Syracuse, Sicily. It is reported by some authors that he visited Egypt and there invented a device now known as Archimedes' screw. This is a pump, still used in many parts of the world. It is highly likely that, when he was a young man, Archimedes studied with the successors of Euclid in Alexandria. Certainly he was completely familiar with the mathematics developed there, but what makes this conjecture much more certain, he knew personally the mathematicians working there and he sent his results to Alexandria with personal messages. He regarded Conon of Samos, one of the mathematicians at Alexandria, both very highly for his abilities as a mathematician and he also regarded him as a close friend.
- tx 237b50dac42af130171773b233954e62690182fd4901a453ad5d11d1d54a8ca3 block 155545 (2012-01-01) contains an exceprt from en.wikipedia.org/wiki/The_Glass_Menagerie
11TomAppearsAtTheTopofTheXXWyM2Bt 11A11eyAfterEachSo1emnBoomXaBp7oy 11ofTheBe111nTheTowerHeXXXXVQaies 11ShakesALitt1eNoisemakeroraeWTgK ... - tx 57bfd63000bbfa6e9a61f7285a4abf9aef91dfcfba4fe0f940b431653eb8068b block 155494 (2011-12-01) is a Lorem ipsum and tx 7961b5ae2f053a16d5c589104f87edfabe80fcae185832ea185e7f0cf06c7747 (2011-12-05) is another one:
11Lorem1psumDo1orSitAmetXXXWAEZ6C 11ConsecteturAdipiscingE1itYQHEPM ...
- tx 1f9606f267cc398356663b14d1a7a3591e3da06572893394c14975a6fc11798f block 155467 (2011-12-01) contains an excerpt from Newton's Principia starting with:
- tx 8ffacbb18f63576fe323cbf2acc6c4c01c86aadf13d8352cfdd39d91916d98c8 block 156164 (2011-12-05) advertises etchablock.com by repeating the following 3 messages 80 times:decoding to:
11EtchABLockDotComGivesYouXZHcYVz 11BLockChain1mmortaLityXXXXYRZD5m 11VisitEtchABLockDotComNowXTbeZZ9More ads can be seen at:etchablock.com gives you blockchain immortaility. Visit etchablock.com now.
- tx 12a8866ea85a8a6838d77cc67ce74ef190a074bc822572f4a82daad00fd980d6 block 156119 (2011-12-05) seems like an alphabet test:
111111111a11111111111b11dC8yHQ 111111111c1111111111111dWctEU9 111111111111e1111111111W7v25m 111111f1111111111111111g11WfG2p8 1111111111111h111111111WdQPXP 11111i111111111111j1111111bL5SyF 11111111k111111111111111XV7PT9D 111L1111111111111111111m111YmYGPJ 1111111111111111n11111U4Rs6D 1111111111111o111111111cLV3wA 11111111p11111111111111qW1RK1A 1111111111111111r1111VRWJZs 111111111111s1111111111VUeyXS 11111111111t11111111111Vq1Wm3 11111111u111111111111111bVpCYE 111111111v11111111111111XoV17A 11111111w1111111111111111YyEFv6 111111111x11111111111111XvZPGp 11111111111y11111111111Y1hDo5 111111111111111zXXXXXXVSn6d5 - tx 31331de21d321766fcac556d7233ad0e3918bc78c7af22b99373569c07d4f30c block 158772 (2011-12-23) has a quick love declaration by a Chinese dude to his Chinese dudesspresumably the man's name is "Ye Chunnan", possible profile: github.com/finway-china
11YechunnanLoveChenchenYeziSsezJQ 11ForeverXXXXXXXXXXXXXXXXXXWcSE4Z - tx bf40e4a1c2546747bc800a085e7145d921a9f402aaf4040c155ff5d0df9cc999 block 161202 (2012-01-08) encodes:
11When1DieBuryMeDeepLayTwoXVEY5jv 11SpeakersAtMyFeetAPairofXXTyrHor 11HeadphonesonMyHeadAndXXXXYUSvnd 11ALwaysPLayTheGratefuLDeadWdq4XoRelated quote mention: www.reddit.com/r/quotes/comments/w51yfg/comment/iwnxk9i/When I die, bury me deep, lay two speakers at my feet, a pair of headphones on my head. Always play the grateful dead.
- tx 3a027fadac6ac2d9cf54480667465ba6ad88b7b3c1de62e1cb34cd06a44243ac block 161267 (2012-01-08) has a birthday wish:
11HappyBirthdayStephenXXXXXZL6eQZ 11HawkingXXXXXXXXXXXXXXXXXXRu9FPeHappy birthday Stephen Hawking
- 1BrianDeeryWasHereCaryNCUSAy1hRCCC on tx c584dd7d3e9ba9776d61ae91f801371e21e434b9d8dab3c81850301433a50fcb block 188657 (2012-06-12) is a check-in by a Brian Deery at Cary, NC, USA. Most likely:
- tx 143a3d7e7599557f9d63e7f224f34d33e9251b2c23c38f95631b3a54de53f024 block 306,204 (2014-06-16) has a Star Wars opening crawl:
1EpisodeiV111111111111111111wbq9i2 1ANewHope1111111111111111111vnYm6D 111111111111111111112xT3273 1itisAPeriodofCiviLWarRebeLyzK2rV 1SpaceshipsStrikingFromA111vh24Fi 1HiddenBaseHaveWonTheirFirstVCugGV 1VictoryAgainstTheEviL111123YSBKF 1GaLacticEmpire1111111111111xsW5HG 1111111111111111111141MmnWZ 1DuringTheBattLeRebeLSpies11ybfhTP 1ManagedToSteaLSecretPLansToxvKf4K 1TheEmpiresULtimateWeapon11zoRcyn 1TheDEATHSTARAnArmoredSpacezUyCHa 1StationWithEnoughPowerTo11vFTWwP 1DestroyAnEntirePLanet1111122KUcy5 111111111111111111114ysyUW1 1PursuedByTheEmpiresSinisterypjWrk 1AgentsPrincessLeiaRacesHomewxuNTT 1AboardHerStarshipCustodian1yhX6zg 1ofTheStoLenPLansThatCan111zCJt3F 1SaveHerPeopLeAndRestore111yULD1y 1FreedomToTheGaLaxy1111111122roNk3 - 1PavedWithGodAndSomeTeensionXudq5X on tx 3e1572ca351d743d7bf627bc844da8f3bdc84eab4a9d27934a8dba30a2e05fe1 block 371894 (2015-08-28) is the largest likely burn that we know of with a single transaction, totalling 1.61803399 BTCIt is unclear what this means exactly and we can't find any pre-existing soruces, but it seems to be a variant of the well known:
Paved With God And Some Teension
- 1NakamotoSatoshiCraigWright8RwLKB appears on two transactions mentioning our friend Craig Steven Wright, the first being tx 2e3207cc93844e2684bdc0bb856c32a6b703dab5b4ba19ed4f06b3fd581b61c3 block 474472 (2016-06-13)A few others include:
Nakamoto Saoshi Craig Wright
- 1FuckRogerVerCraigWrightJihanBGMX3 (2018-02-08)
- 1CraigWrightisAFrausterCuntwgASwJ (2019-06-02)
Craig Wright is a fraudster cunt
- 1FuckYouGraigWrightxSatoshiXc6ppN (2020-06-24)
Fuck Craig Wright
Related:
- www.reddit.com/r/Buttcoin/comments/3kqdjv/a_list_of_bitcoin_addresses_used_to_intentionally/ A list of bitcoin addresses used to intentionally burn bitcoin (2015-09-13). Their list is not based solely on base58 images, e.g. 1A1zP1eP5QGefi2DMPTfTL5SLmv7DivfNa from the Genesis block is present. Also their ordering is unclear, it's neither stricly chronological nor by value. But it is a good list however.
- github.com/BranndonWork/bitcoin-bulk-balance-check/blob/master/burned.csv
- bitcointalk.org/index.php?topic=84569 Vanity Pool - vanity address generator pool
- bitcointalk.org/index.php?topic=85935 Coins sent to the great wallet in the sky (2012-06-07)
- bitcointalk.org/index.php?topic=90982.285 Rare address hall of fame (2015)
- bitcointalk.org/index.php?topic=917913.0 Burn Baby Burn! - Compiling all Bitcoin burn addresses by fairglu (2015)
- bitcointalk.org/index.php?topic=553449.0 Longest most impressive VANITY (2014)
- bitcointalk.org/index.php?topic=558604.0 Custom Bitcoin address? (2014)
- medium.com/@westkate37/burned-bitcoins-d9b15b3699d6
- www.bitcoinwhoswho.com/blog/2018/12/29/2-btc-burned-in-2018/
- bitcoin.stackexchange.com/questions/70241/whats-with-this-address-1111111111111111111114olvt2/125931#125931
- bitcoin.stackexchange.com/questions/49625/whats-the-point-of-bitcoin-eaters mentions in particular
1BitcoinEaterAddressDontSendf59kuE - kf106.medium.com/interesting-addresses-on-the-bitcoin-blockchain-e0956f06ec01 has some interesting monetary ones, not inscriptions
- www.bitcoinwhoswho.com/blog/2016/06/01/the-7-most-incredible-bitcoin-addresses/ The 7 Most Incredible Bitcoin Addresses (2016)
Cool data embedded in the Bitcoin blockchain The largest transactions in the Bitcoin Blockchain by  Ciro Santilli 40 Updated 2025-07-16
Ciro Santilli 40 Updated 2025-07-16
- bb41a757f405890fb0f5856228e23b715702d714d59bf2b1feb70d8b2b4e3e08 999,657 bytes. Joins a bunch of tiny inputs into a single output
- 623463a2a8a949e0590ffe6b2fd3e4e1028b2b99c747e82e899da4485eb0b6be and 5143cf232576ae53e8991ca389334563f14ea7a7c507a3e081fbef2538c84f6e both have 3,075 outputs of 1 satoshi each and a single input. We were not able to identify any meaningful data in it,
filejust saysdata, and there aren't long ASCII strings. However, the outputs were unspent as of 2021, which suggests that they might actually be data.
Analysis of some of them follows.
- dd9f6bbf80ab36b722ca95d93268667a3ea6938288e0d4cf0e7d2e28a7a91ab3 has 13107 with payload 256KB in size, but some of them at least have been spent: www.blockchain.com/btc/tx/dd9f6bbf80ab36b722ca95d93268667a3ea6938288e0d4cf0e7d2e28a7a91ab3 therefore it's not data.
filesays their payload is a DOS executable, but it must be a coincidence
Matrix congruence can be seen as the change of basis of a bilinear form by  Ciro Santilli 40 Updated 2025-07-16
Ciro Santilli 40 Updated 2025-07-16
From effect of a change of basis on the matrix of a bilinear form, remember that a change of basis modifies the matrix representation of a bilinear form as:
So, by taking , we understand that two matrices being congruent means that they can both correspond to the same bilinear form in different bases.
Some nice insights at: Robert Noyce: The Man Behind the Microchip by Leslie Berlin (2006).
They are the finite projective special linear groups.
This was the first infinite family of simple groups discovered after the simple cyclic groups and alternating groups. The first case discovered was by Galois. You should understand that one first.
Used to explain the black-body radiation experiment.
Published as: On the Theory of the Energy Distribution Law of the Normal Spectrum by Max Planck (1900).
The Quantum Story by Jim Baggott (2011) page 9 mentions that Planck apparently immediately recognized that Planck constant was a new fundamental physical constant, and could have potential applications in the definition of the system of units (TODO where was that published):This was a visionary insight, and was finally realized in the 2019 redefinition of the SI base units.
Planck wrote that the constants offered: 'the possibility of establishing units of length, mass, time and temperature which are independent of specific bodies or materials and which necessarily maintain their meaning for all time and for all civilizations, even those which are extraterrestrial and nonhuman, constants which therefore can be called "fundamental physical units of measurement".'
TODO how can it be derived from theoretical principles alone? There is one derivation at; en.wikipedia.org/wiki/Planck%27s_law#Derivation but it does not seem to mention the Schrödinger equation at all.
Using de novo DNA synthesis to synthesize entire Chromosomes.
There are unlisted articles, also show them or only show them.




:max_bytes(150000):strip_icc()/star-wars-crawl-02-d8ed15dbd1fe4127858c3e22593988be.jpg)
