Some criticisms:
Ciro Santilli invented this term, derived from "hardware in the loop" to refer to simulations in which both the brain and the body and physical world of organism models are modelled.
E.g. just imagine running:
Likely implies whole brain emulation and therefore AGI.
It's like the bootloader stage of biology! It's weird and magic and important: Section "Molecular biology feels like systems programming".
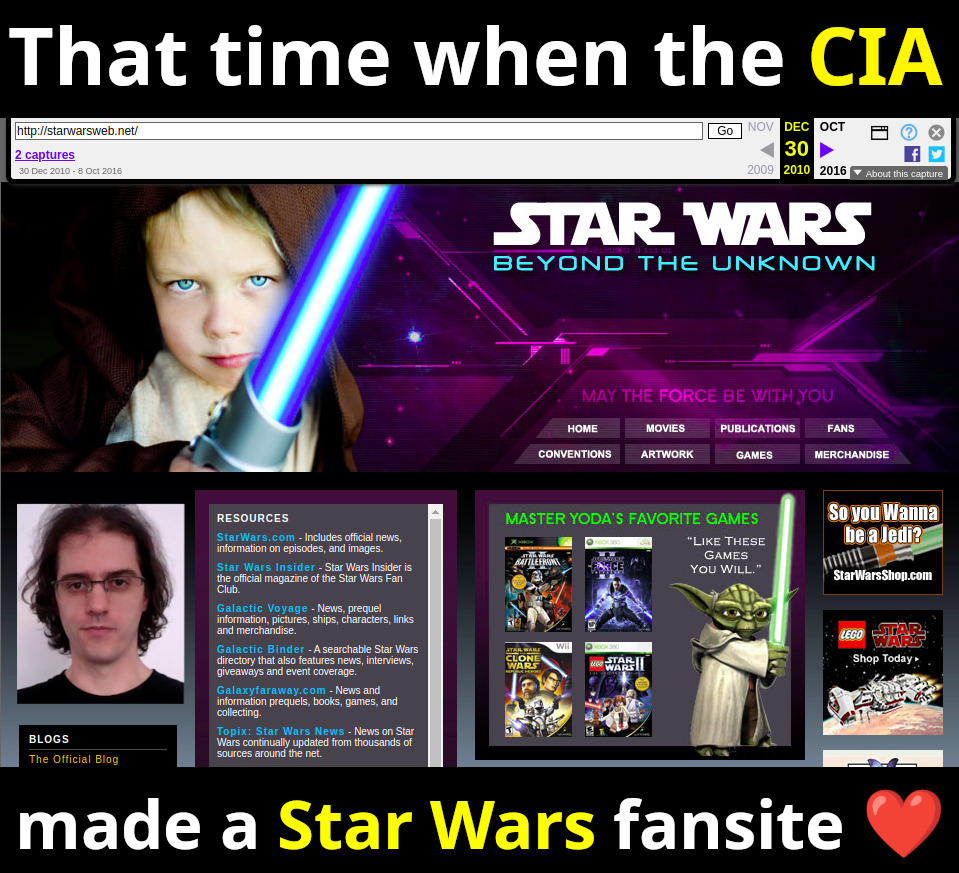
This article is about covert agent communication channel websites used by the CIA in many countries from the mid 2000s until the early 2010s, when they were uncovered by counter intelligence of some of the targeted countries, notably Iran and China, circa 2010-2013.
This article uses publicly available information to publicly disclose for the first time a few hundred of what we feel are extremely likely candidate sites of the network. The starting point for this research was the September 2022 Reuters article "America’s Throwaway Spies" which for the first time gave nine example websites, and their analyst from Citizenlabs claims to have found 885 websites in total, but did not publicly disclose them. Starting from only the nine disclosed websites, we were then able to find a few hundred websites that share so many similarities with them, i.e. a common fingerprint, that we believe makes them beyond reasonable doubt part of the same network.
If you enjoy this article, consider dropping some Monero at: 4A1KK4uyLQX7EBgN7uFgUeGt6PPksi91e87xobNq7bT2j4V6LqZHKnkGJTUuCC7TjDNnKpxDd8b9DeNBpSxim8wpSczQvzf so I can waste it on my foolish attempts to improve higher education. Other sponsorship methods: Section "Sponsor Ciro Santilli's work on OurBigBook.com".
The discovery of these websites by Iranian and Chinese counterintelligence led to the imprisonment and execution of several assets in those countries, and subsequent shutdown of the channel by the CIA when they noticed that things had gone wrong. This is likely a Wikipedia page that talks about the disastrous outcome of the websites being found out: 2010–2012 killing of CIA sources in China, although it contained no mention of websites before Ciro Santilli edited it in.
Of particular interest is that based on their language and content, certain of the websites seem to have targeted other democracies such as Germany, France, Spain and Brazil.
If anyone can find others websites, or has better techniques feel free to contact Ciro Santilli at: Section "How to contact Ciro Santilli". Contributions will be clearly attributed if desired. Some of the techniques used so far have been very heuristic, and that added to the limited amount of data makes it almost certain that some websites have been missed. Broadly speaking, there are two types of contributions that would be possible:
- finding new IP ranges: harder and more exiting, and potentially requires more intelligence
- better IP to domain name databases to fill in known gaps in existing IP ranges
The fact that citizenlabs reported exactly 885 websites being found makes it feel like they might have found find a better fingerprint which we have not managed to find yet. We have not yet had to pay for our data. If someone wants to donate to the research, some ideas include:
- dump $400 on WhoisXMLAPI to dump whois history of all known hits and search for other matches. Small discoveries were made like this in the past and we'd expect a few more to be left. We don't expect huge breakthroughs from this, but at only $400 it is not so bad
- dump a lot more ($15k+? needs confirmation as opaque pricing) on DomainTools. We are not certain that they have any superior data since there is no free trial of any kind, but it would be interesting to test the quality of the data they acquired from Farsight DNSDB if you are really loaded
Disclaimers:
- the network fell in 2013, followed by fully public disclosures in 2018 and 2022, so we believe that the benefit of giving the public this broader historic understanding outweighs the risks that agents could be found so many years later by sloppy secret services
- Ciro Santilli's political bias is strongly pro-democracy and anti-dictatorship, but with a good pinch of skepticism about the morality US foreign policy in the last century
CLI program implementing Universal Chess Interface: www.reddit.com/r/ComputerChess/comments/b6rdez/commandline_options_for_stockfish/
How to actually play against it: chess.stackexchange.com/questions/4353/how-to-install-stockfish-on-ubuntu So hard!
The user friendly Chess UI! Exactly what you would expect from a GNOME Project package. But also packs some punch via the Universal Chess Interface, e.g. Stockfish just works.
Both chess engine and a CLI chess UI. As an engine it is likely irrelevant compared to Stockfish as of 2020. TODO: does the UI support Universal Chess Interface?
Cool project history though. Started before the GNU Project itself, and became one of the first packages.
Advanced. Not beginner friendly, very clunky.
To be fair, this is one of the least worse ones.
The cool thing about this notation is that is showed to Ciro Santilli that there is more state to a chess game than just the board itself! Notably:plus some other boring draw rules counters.
- whose move it is next
- castling availability
- en passant availability
There are unlisted articles, also show them or only show them.