100 Greatest Discoveries by the Discovery Channel (2004-2005) Updated 2025-07-16
Hosted by Bill Nye.
Physics topics:
- Galileo: objects of different masses fall at the same speed, hammer and feather experiment
- Newton: gravity, linking locally observed falls and the movement of celestial bodies
- TODO a few more
- superconductivity, talk only at Fermilab accelerator, no re-enactment even...
- quark, interview with Murray Gell-Mann, mentions it was "an off-beat field, one wasn't encouraged to work on that". High level blablabla obviously.
- fundamental interactions, notably weak interaction and strong interaction, interview with Michio Kaku. When asked "How do we know that the weak force is there?" the answer is: "We observe radioactive decay with a Geiger counter". Oh, come on!
biology topics:
- Leeuwenhoek microscope and the discovery of microorganisms, and how pond water is not dead, but teeming with life. No sample of course.
- 1831 Robert Brown cell nucleus in plants, and later Theodor Schwann in tadpoles. This prepared the path for the idea that "all cells come from other cells", and the there seemed to be an unifying theme to all life: the precursor to DNA discoveries. Re-enactment, yay.
- 1971 Carl Woese and the discovery of archaea
Genetics:
- Mendel. Reenactment.
- 1909 Thomas Hunt Morgan with Drosophila melanogaster. Reenactment. Genes are in Chromosomes. He observed that a trait was linked to sex, and it was already known that sex was related to chromosomes.
- 1935 George Beadle and the one gene one enzyme hypothesis by shooting X-rays at bread mold
- 1942 Barbara McClintock, at Cold Spring Harbor Laboratory
- 1952 Hershey–Chase experiment. Determined that DNA is what transmits genetic information, not protein, by radioactive labelling both protein and DNA in two sets of bacteriophages. They observed that only the DNA radioactive material was passed forward.
- Crick Watson
- messenger RNA, no specific scientist, too many people worked on it, done partially with bacteriophage experiments
- 1968 Nirenberg genetic code
- 1972 Hamilton O. Smith and the discovery of restriction enzymes by observing that they were part of anti bacteriophage immune-system present in bacteria
- alternative splicing
- RNA interference
- Human Genome Project, interview with Craig Venter.
Medicine:
- blood circulation
- anesthesia
- X-ray
- germ theory of disease, with examples from Ignaz Semmelweis and Pasteur
- 1796 Edward Jenner discovery of vaccination by noticing that cowpox cowpox infected subjects were immune
- vitamin by observing scurvy and beriberi in sailors, confirmed by Frederick Gowland Hopkins on mice experiments
- Fleming, Florey and Chain and the discovery of penicillin
- Prontosil
- diabetes and insulin
Animation of molecular biology processes Updated 2025-07-16
Nothing makes the fact that your life is an illusion clearer than animations of molecular biology processes. You just have no idea what is going on inside your own body right now!
And yet, we live, oblivious to all of it.
Amazing creators:
Electron transport chain by HarvardX (2017)
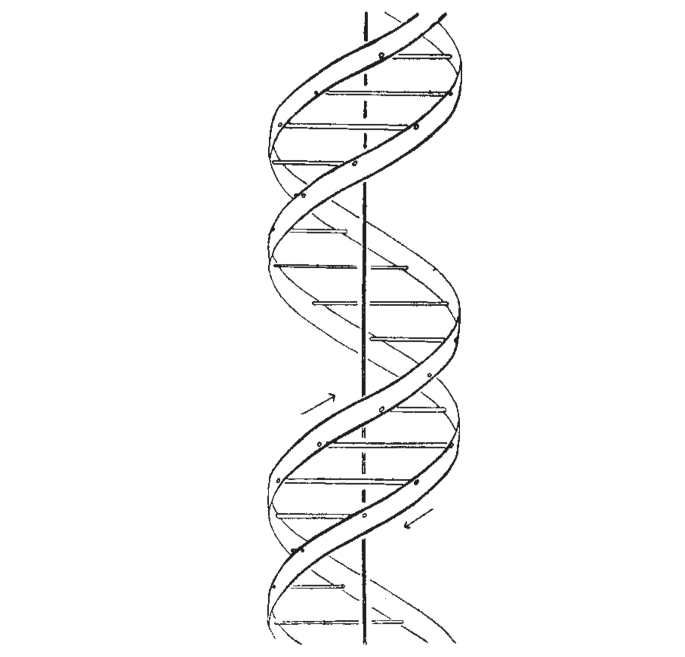
Source. This actually explains how mitochondrions use sugar derivatives and oxygen to transform ADP into ATP. A Structure for Deoxyribose Nucleic Acid Created 2025-06-12 Updated 2025-07-16
Starting line:The Eighth Day of Creation explains the "salt" part as that was the usual way to prepare DNA for X-ray crystallography, where something binds with the phosphate groups of DNA
We wish to suggest a structure for the salt of deoxyribose nucleic acid (D.N.A,). This structure has novel features which are of considerable biological interest.
The paper then shoots down other previously devised helical structures, notably some containing 3 strands or phosphate on the inside.
Then they briefly describe their structure, and promise more details on future articles. This was mostly a short one-page priority note.
Then they drop their shell bomb conclusion:
It has not es~aped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.
Both Wilkins and Rosalind Franklin are acknowledged at the end.
DNA microarray Updated 2025-07-16
Can be seen as a cheap form of DNA sequencing that only test for a few hits. Some major applications:
- gene expression profiling
- single-nucleotide polymorphism: specificity is high enough to detect snips
E. Coli Whole Cell Model by Covert Lab Updated 2025-07-16
github.com/CovertLab/WholeCellEcoliRelease is a whole cell simulation model created by Covert Lab and other collaborators.
The project is written in Python, hurray!
But according to te README, it seems to be the use a code drop model with on-request access to master. Ciro Santilli asked at rationale on GitHub discussion, and they confirmed as expected that it is to:
- to prevent their publication ideas from being stolen. Who would steal publication ideas with public proof in an issue tracker without crediting original authors? Academia is broken. Academia should be the most open form of knowledge sharing. But instead we get this silly competition for publication points.
- to prevent noise from non-collaborators. But they only get like 2 issues as year on such a meganiche subject... Did you know that you can ignore people, and even block them if they are particularly annoying? Much more likely is that no one will every hear about your project and that it will die with its last graduate student slave.
The project is a followup to the earlier M. genitalium whole cell model by Covert lab which modelled Mycoplasma genitalium. E. Coli has 8x more genes (500 vs 4k), but it the undisputed bacterial model organism and as such has been studied much more thoroughly. It also reproduces faster than Mycoplasma (20 minutes vs a few hours), which is a huge advantages for validation/exploratory experiments.
The project has a partial dependency on the proprietary optimization software CPLEX which is freeware, for students, not sure what it is used for exactly, from the comment in the
requirements.txt the dependency is only partial.This project makes Ciro Santilli think of the E. Coli as an optimization problem. Given such external nutrient/temperature condition, which DNA sequence makes the cell grow the fastest? Balancing metabolites feels like designing a Factorio speedrun.
There is one major thing missing thing in the current model: promoters/transcription factor interactions are not modelled due to lack/low quality of experimental data: github.com/CovertLab/WholeCellEcoliRelease/issues/21. They just have a magic direct "transcription factor to gene" relationship, encoded at reconstruction/ecoli/flat/foldChanges.tsv in terms of type "if this is present, such protein is expressed 10x more". Transcription units are not implemented at all it appears.
Everything in this section refers to version 7e4cc9e57de76752df0f4e32eca95fb653ea64e4, the code drop from November 2020, and was tested on Ubuntu 21.04 with a docker install of
docker.pkg.github.com/covertlab/wholecellecolirelease/wcm-full with image id 502c3e604265, unless otherwise noted. E. Coli Whole Cell Model by Covert Lab Mass fraction summary plot analysis Created 2024-12-04 Updated 2025-07-16
Let's look into a sample plot,
out/manual/plotOut/svg_plots/massFractionSummary.svg, and try to understand as much as we can about what it means and how it was generated.This plot contains how much of each type of mass is present in all cells. Since we simulated just one cell, it will be the same as the results for that cell.
We can see that all of them grow more or less linearly, perhaps as the start of an exponential. We can see that all of them grow more or less linearly, perhaps as the start of an exponential. We can see that all of them grow more or less linearly, perhaps as the start of an exponential.which must correspond to the different
By grepping the title "Cell mass fractions" in the source code, we see the files:
models/ecoli/analysis/cohort/massFractionSummary.py
models/ecoli/analysis/multigen/massFractionSummary.py
models/ecoli/analysis/variant/massFractionSummary.pymassFractionSummary plots throughout different levels of the hierarchy.By reading
models/ecoli/analysis/variant/massFractionSummary.py a little bit, we see that:- the plotting is done with Matplotlib, hurray
- it is reading its data from files under
./out/manual/wildtype_000000/000000/generation_000000/000000/simOut/Mass/, more precisely./out/manual/wildtype_000000/000000/generation_000000/000000/simOut/Mass/columns/<column-name>/data. They are binary files however.Looking at the source forwholecell/io/tablereader.pyshows that those are just a standard NumPy serialization mechanism. Maybe they should have used the Hierarchical Data Format instead.We can also take this opportunity to try and find where the data is coming from.Massfrom the./out/manual/wildtype_000000/000000/generation_000000/000000/simOut/Mass/looks like an ID, so wegrepthat and we reachmodels/ecoli/listeners/mass.py.From this we understand that all data that is to be saved from a simulation must be coming from listeners: likely nothing, or not much, is dumped by default, because otherwise it would take up too much disk space. You have to explicitly say what it is that you want to save via a listener that acts on each time step.
More plot types will be explored at time series run variant, where we will contrast two runs with different growth mediums.
E. Coli Whole Cell Model by Covert Lab Output overview Updated 2025-07-16
Run output is placed under
out/:Some of the output data is stored as
.cpickle files. To observe those files, you need the original Python classes, and therefore you have to be inside Docker, from the host it won't work.We can list all the plots that have been produced under Plots are also available in SVG and PDF formats, e.g.:
out/ withfind -name '*.png'The output directory has a hierarchical structure of type:where:
./out/manual/wildtype_000000/000000/generation_000000/000000/wildtype_000000: variant conditions.wildtypeis a human readable label, and000000is an index amongst the possiblewildtypeconditions. For example, we can have different simulations with different nutrients, or different DNA sequences. An example of this is shown at run variants.000000: initial random seed for the initial cell, likely fed to NumPy'snp.random.seedgenereation_000000: this will increase with generations if we simulate multiple cells, which is supported by the model000000: this will presumably contain the cell index within a generation
We also understand that some of the top level directories contain summaries over all cells, e.g. the
massFractionSummary.pdf plot exists at several levels of the hierarchy:./out/manual/plotOut/massFractionSummary.pdf
./out/manual/wildtype_000000/plotOut/massFractionSummary.pdf
./out/manual/wildtype_000000/000000/plotOut/massFractionSummary.pdf
./out/manual/wildtype_000000/000000/generation_000000/000000/plotOut/massFractionSummary.pdfEach of thoes four levels of
plotOut is generated by a different one of the analysis scripts:./out/manual/plotOut: generated bypython runscripts/manual/analysisVariant.py. Contains comparisons of different variant conditions. We confirm this by looking at the results of run variants../out/manual/wildtype_000000/plotOut: generated bypython runscripts/manual/analysisCohort.py --variant_index 0. TODO not sure how to differentiate between two different labels e.g.wildtype_000000andsomethingElse_000000. If-vis not given, a it just picks the first one alphabetically. TODO not sure how to automatically generate all of those plots without inspecting the directories../out/manual/wildtype_000000/000000/plotOut: generated bypython runscripts/manual/analysisMultigen.py --variant_index 0 --seed 0./out/manual/wildtype_000000/000000/generation_000000/000000/plotOut: generated bypython runscripts/manual/analysisSingle.py --variant_index 0 --seed 0 --generation 0 --daughter 0. Contains information about a single specific cell.
E. Coli Whole Cell Model by Covert Lab Source code overview Updated 2025-07-16
Let's try to understand some interesting looking, with a special focus on our understanding of the tiny E. Coli K-12 MG1655 operon thrLABC part of the metabolism, which we have well understood at Section "E. Coli K-12 MG1655 operon thrLABC".
reconstruction/ecoli/flat/compartments.tsvcontains cellular compartment information:"abbrev" "id" "n" "CCO-BAC-NUCLEOID" "j" "CCO-CELL-PROJECTION" "w" "CCO-CW-BAC-NEG" "c" "CCO-CYTOSOL" "e" "CCO-EXTRACELLULAR" "m" "CCO-MEMBRANE" "o" "CCO-OUTER-MEM" "p" "CCO-PERI-BAC" "l" "CCO-PILUS" "i" "CCO-PM-BAC-NEG"CCO: "Celular COmpartment"BAC-NUCLEOID: nucleoidCELL-PROJECTION: cell projectionCW-BAC-NEG: TODO confirm: cell wall (of a Gram-negative bacteria)CYTOSOL: cytosolEXTRACELLULAR: outside the cellMEMBRANE: cell membraneOUTER-MEM: bacterial outer membranePERI-BAC: periplasmPILUS: pilusPM-BAC-NEG: TODO: plasma membrane, but that is the same as cell membrane no?
reconstruction/ecoli/flat/promoters.tsvcontains promoter information. Simple file, sample lines:corresponds to E. Coli K-12 MG1655 promoter thrLp, which starts as position 148."position" "direction" "id" "name" 148 "+" "PM00249" "thrLp"reconstruction/ecoli/flat/proteins.tsvcontains protein information. Sample line corresponding to e. Coli K-12 MG1655 gene thrA:so we understand that:"aaCount" "name" "seq" "comments" "codingRnaSeq" "mw" "location" "rnaId" "id" "geneId" [91, 46, 38, 44, 12, 53, 30, 63, 14, 46, 89, 34, 23, 30, 29, 51, 34, 4, 20, 0, 69] "ThrA" "MRVL..." "Location information from Ecocyc dump." "AUGCGAGUGUUG..." [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 89103.51099999998, 0.0, 0.0, 0.0, 0.0] ["c"] "EG10998_RNA" "ASPKINIHOMOSERDEHYDROGI-MONOMER" "EG10998"aaCount: amino acid count, how many of each of the 20 proteinogenic amino acid are thereseq: full sequence, using the single letter abbreviation of the proteinogenic amino acidsmw; molecular weight? The 11 components appear to be given atreconstruction/ecoli/flat/scripts/unifyBulkFiles.py:so they simply classify the weight? Presumably this exists for complexes that have multiple classes?molecular_weight_keys = [ '23srRNA', '16srRNA', '5srRNA', 'tRNA', 'mRNA', 'miscRNA', 'protein', 'metabolite', 'water', 'DNA', 'RNA' # nonspecific RNA ]23srRNA,16srRNA,5srRNAare the three structural RNAs present in the ribosome: 23S ribosomal RNA, 16S ribosomal RNA, 5S ribosomal RNA, all others are obvious:- tRNA
- mRNA
- protein. This is the seventh class, and this enzyme only contains mass in this class as expected.
- metabolite
- water
- DNA
- RNA: TODO
rnavsmiscRNA
location: cell compartment where the protein is present,cdefined atreconstruction/ecoli/flat/compartments.tsvas cytoplasm, as expected for something that will make an amino acid
reconstruction/ecoli/flat/rnas.tsv: TODO vstranscriptionUnits.tsv. Sample lines:"halfLife" "name" "seq" "type" "modifiedForms" "monomerId" "comments" "mw" "location" "ntCount" "id" "geneId" "microarray expression" 174.0 "ThrA [RNA]" "AUGCGAGUGUUG..." "mRNA" [] "ASPKINIHOMOSERDEHYDROGI-MONOMER" "" [0.0, 0.0, 0.0, 0.0, 790935.00399999996, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0] ["c"] [553, 615, 692, 603] "EG10998_RNA" "EG10998" 0.0005264904halfLife: half-lifemw: molecular weight, same as inreconstruction/ecoli/flat/proteins.tsv. This molecule only have weight in themRNAclass, as expected, as it just codes for a proteinlocation: same as inreconstruction/ecoli/flat/proteins.tsvntCount: nucleotide count for each of the ATGCmicroarray expression: presumably refers to DNA microarray for gene expression profiling, but what measure exactly?
reconstruction/ecoli/flat/sequence.fasta: FASTA DNA sequence, first two lines:>E. coli K-12 MG1655 U00096.2 (1 to 4639675 = 4639675 bp) AGCTTTTCATTCTGACTGCAACGGGCAATATGTCTCTGTGTGGATTAAAAAAAGAGTGTCTGATAGCAGCTTCTGreconstruction/ecoli/flat/transcriptionUnits.tsv: transcription units. We can observe for example the two different transcription units of the E. Coli K-12 MG1655 operon thrLABC in the lines:"expression_rate" "direction" "right" "terminator_id" "name" "promoter_id" "degradation_rate" "id" "gene_id" "left" 0.0 "f" 310 ["TERM0-1059"] "thrL" "PM00249" 0.198905992329492 "TU0-42486" ["EG11277"] 148 657.057317358791 "f" 5022 ["TERM_WC-2174"] "thrLABC" "PM00249" 0.231049060186648 "TU00178" ["EG10998", "EG10999", "EG11000", "EG11277"] 148promoter_id: matches promoter id inreconstruction/ecoli/flat/promoters.tsvgene_id: matches id inreconstruction/ecoli/flat/genes.tsvid: matches exactly those used in BioCyc, which is quite nice, might be more or less standardized:
reconstruction/ecoli/flat/genes.tsv"length" "name" "seq" "rnaId" "coordinate" "direction" "symbol" "type" "id" "monomerId" 66 "thr operon leader peptide" "ATGAAACGCATT..." "EG11277_RNA" 189 "+" "thrL" "mRNA" "EG11277" "EG11277-MONOMER" 2463 "ThrA" "ATGCGAGTGTTG" "EG10998_RNA" 336 "+" "thrA" "mRNA" "EG10998" "ASPKINIHOMOSERDEHYDROGI-MONOMER"reconstruction/ecoli/flat/metabolites.tsvcontains metabolite information. Sample lines:In the case of the enzyme thrA, one of the two reactions it catalyzes is "L-aspartate 4-semialdehyde" into "Homoserine"."id" "mw7.2" "location" "HOMO-SER" 119.12 ["n", "j", "w", "c", "e", "m", "o", "p", "l", "i"] "L-ASPARTATE-SEMIALDEHYDE" 117.104 ["n", "j", "w", "c", "e", "m", "o", "p", "l", "i"]Starting from the enzyme page: biocyc.org/gene?orgid=ECOLI&id=EG10998 we reach the reaction page: biocyc.org/ECOLI/NEW-IMAGE?type=REACTION&object=HOMOSERDEHYDROG-RXN which has reaction IDHOMOSERDEHYDROG-RXN, and that page which clarifies the IDs:so these are the compounds that we care about.- biocyc.org/compound?orgid=ECOLI&id=L-ASPARTATE-SEMIALDEHYDE: "L-aspartate 4-semialdehyde" has ID
L-ASPARTATE-SEMIALDEHYDE - biocyc.org/compound?orgid=ECOLI&id=HOMO-SER: "Homoserine" has ID
HOMO-SER
- biocyc.org/compound?orgid=ECOLI&id=L-ASPARTATE-SEMIALDEHYDE: "L-aspartate 4-semialdehyde" has ID
reconstruction/ecoli/flat/reactions.tsvcontains chemical reaction information. Sample lines:"reaction id" "stoichiometry" "is reversible" "catalyzed by" "HOMOSERDEHYDROG-RXN-HOMO-SER/NAD//L-ASPARTATE-SEMIALDEHYDE/NADH/PROTON.51." {"NADH[c]": -1, "PROTON[c]": -1, "HOMO-SER[c]": 1, "L-ASPARTATE-SEMIALDEHYDE[c]": -1, "NAD[c]": 1} false ["ASPKINIIHOMOSERDEHYDROGII-CPLX", "ASPKINIHOMOSERDEHYDROGI-CPLX"] "HOMOSERDEHYDROG-RXN-HOMO-SER/NADP//L-ASPARTATE-SEMIALDEHYDE/NADPH/PROTON.53." {"NADPH[c]": -1, "NADP[c]": 1, "PROTON[c]": -1, "L-ASPARTATE-SEMIALDEHYDE[c]": -1, "HOMO-SER[c]": 1 false ["ASPKINIIHOMOSERDEHYDROGII-CPLX", "ASPKINIHOMOSERDEHYDROGI-CPLX"]catalized by: here we seeASPKINIHOMOSERDEHYDROGI-CPLX, which we can guess is a protein complex made out ofASPKINIHOMOSERDEHYDROGI-MONOMER, which is the ID for thethrAwe care about! This is confirmed incomplexationReactions.tsv.
reconstruction/ecoli/flat/complexationReactions.tsvcontains information about chemical reactions that produce protein complexes:The"process" "stoichiometry" "id" "dir" "complexation" [ { "molecule": "ASPKINIHOMOSERDEHYDROGI-CPLX", "coeff": 1, "type": "proteincomplex", "location": "c", "form": "mature" }, { "molecule": "ASPKINIHOMOSERDEHYDROGI-MONOMER", "coeff": -4, "type": "proteinmonomer", "location": "c", "form": "mature" } ] "ASPKINIHOMOSERDEHYDROGI-CPLX_RXN" 1coeffis how many monomers need to get together for form the final complex. This can be seen from the Summary section of ecocyc.org/gene?orgid=ECOLI&id=ASPKINIHOMOSERDEHYDROGI-MONOMER:Fantastic literature summary! Can't find that in database form there however.Aspartate kinase I / homoserine dehydrogenase I comprises a dimer of ThrA dimers. Although the dimeric form is catalytically active, the binding equilibrium dramatically favors the tetrameric form. The aspartate kinase and homoserine dehydrogenase activities of each ThrA monomer are catalyzed by independent domains connected by a linker region.
reconstruction/ecoli/flat/proteinComplexes.tsvcontains protein complex information:"name" "comments" "mw" "location" "reactionId" "id" "aspartate kinase / homoserine dehydrogenase" "" [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 356414.04399999994, 0.0, 0.0, 0.0, 0.0] ["c"] "ASPKINIHOMOSERDEHYDROGI-CPLX_RXN" "ASPKINIHOMOSERDEHYDROGI-CPLX"reconstruction/ecoli/flat/protein_half_lives.tsvcontains the half-life of proteins. Very few proteins are listed however for some reason.reconstruction/ecoli/flat/tfIds.csv: transcription factors information:"TF" "geneId" "oneComponentId" "twoComponentId" "nonMetaboliteBindingId" "activeId" "notes" "arcA" "EG10061" "PHOSPHO-ARCA" "PHOSPHO-ARCA" "fnr" "EG10325" "FNR-4FE-4S-CPLX" "FNR-4FE-4S-CPLX" "dksA" "EG10230"
Human mitochondrion Updated 2025-07-16
DNA stuff at: human mtDNA.
Molecular biology feels like systems programming Updated 2025-07-16
Whenever Ciro Santilli learns about molecular biology, he can't help but to feel that it feels like programming, and notably systems programming and computer hardware design.
In some sense, the comparison is obvious: DNA is clearly a programmable medium like any assembly language, but still, systems programming did give Ciro some further feelings.
- The most important analogy perhaps is observability, or more precisely the lack of it. For the computer, this is described at: The lower level you go into a computer, the harder it is to observe things.And then, when Ciro started learning a bit about biology techniques, he started to feel the exact same thing.For example when he played with E. Coli Whole Cell Model by Covert Lab, the main thing Ciro felt was: it is going to be hard to verify any of this data, because it is hard/impossible to know the concentration of each element in a cell as a function of time.More generally of course, this is exactly why making any biology discovery is so hard: we can't easily see what's going on inside the cell, and have to resort to indirect ways of doing so..This exact idea was highlighted by I should have loved biology by James Somers:
For a computer scientist, a biologist's methods can seem insane; the trouble comes from the fact that cells are too small, too numerous, too complex to analyze the way a programmer would, say in a step-by-step debugger.
And then just like in software, some of the methods biologists use to overcome the lack of visibility have direct software analogues:- add instrumentation to cells, e.g. GFP tagging comes to mind
- emulation, e.g. E. Coli Whole Cell Model by Covert Lab
- The boot process is another one. E.g. in x86 the way that you start in 16-bit mode, largely compatible into the 70's, then move to 32-bit and finally 64, does feel a lot the way a earlier stages of embryo development looks more and more like more ancient animals.
Ciro likes to think that maybe that is why a hardcore systems programmer like Bert Hubert got into molecular biology.
Some other people who mention similar things:
- I should have loved biology by James Somers highlights the computer abstraction layer analogy between the two:
Molecular biology technologies Updated 2025-07-16
As of 2019, the silicon industry is ending, and molecular biology technology is one of the most promising and growing field of engineering.
Ciro Santilli is especially excited about DNA-related technologies, because DNA is the centerpiece of biology, and it is programmable.
First, during the 2000's, the cost of DNA sequencing fell to about 1000 USD per genome in the end of the 2010's: Figure 2. "Cost per genome vs Moore's law from 2000 to 2019", largely due to "Illumina's" technology.
The medical consequences of this revolution are still trickling down towards medical applications of 2019, inevitably, but somewhat slowly due to tight privacy control of medical records.
Ciro Santilli predicts that when the 100 dollar mark is reached, every person of the First world will have their genome sequenced, and then medical applications will be closer at hand than ever.
But even 100 dollars is not enough. Sequencing power is like computing power: humankind can never have enough. Sequencing is not a one per person thing. For example, as of 2019 tumors are already being sequenced to help understand and treat them, and scientists/doctors will sequence as many tumor cells as budget allows.
Then, in the 2010's, CRISPR/Cas9 gene editing started opening up the way to actually modifying the genome that we could now see through sequencing.
What's next?
Ciro believes that the next step in the revolution could be could be: de novo DNA synthesis.
This technology could be the key to the one of the ultimate dream of biologists: cheap programmable biology with push-button organism bootstrap!
Just imagine this: at the comfort of your own garage, you take some model organism of interest, maybe start humble with Escherichia coli. Then you modify its DNA to your liking, and upload it to a 3D printer sized machine on your workbench, which automatically synthesizes the DNA, and injects into a bootstrapped cell.
You then make experiments to check if the modified cell achieves your desired new properties, e.g. production of some protein, and if not reiterate, just like a software engineer.
Of course, even if we were able to do the bootstrap, the debugging process then becomes key, as visibility is the key limitation of biology, maybe we need other cheap technologies to come in at that point.
This a place point we see the beauty of evolution the brightest: evolution does not require observability. But it also implies that if your changes to the organism make it less fit, then your mutation will also likely be lost. This has to be one of the considerations done when designing your organism.
Other cool topic include:
- computational biology: simulations of cell metabolism, protein and small molecule, including computational protein folding and chemical reactions. This is basically the simulation part of omics.If we could only simulate those, we would basically "solve molecular biology". Just imagine, instead of experimenting for a hole year, the 2021 Nobel Prize in Physiology and Medicine could have been won from a few hours on a supercomputer to determine which protein had the desired properties, using just DNA sequencing as a starting point!
- microscopy: crystallography, cryoEM
- analytical chemistry: mass spectroscopy, single cell analysis (Single-cell RNA sequencing)
Ciro is sad that by the time he dies, humanity won't have understood the human brain, maybe not even a measly Escherichia coli... Heck, even key molecular biology events are not yet fully understood, see e.g. transcription regulation.
One of the most exciting aspects of molecular biology technologies is their relatively low entry cost, compared for example to other areas such as fusion energy and quantum computing.
OpenWorm Updated 2025-07-16
High level simulation only, no way to get from DNA to worm! :-) Includes:
- nervous system
- muscle system
OpenWorm Sibernetic demo by Mike Vella (2013)
Source. Sibernetic adds a fluid dynamics solver for brain-in-the-loop simulation of C. elegans. How to use an Oxford Nanopore MinION to extract DNA from river water and determine which bacteria live in it Overview of the experiment Updated 2025-07-16
For those that know biology and just want to do the thing, see: Section "Protocols used".
The PuntSeq team uses an Oxford Nanopore MinION DNA sequencer made by Oxford Nanopore Technologies to sequence the 16S region of bacterial DNA, which is about 1500 nucleotides long.
This kind of "decode everything from the sample to see what species are present approach" is called "metagenomics".
This is how the MinION looks like: Figure 1. "Oxford Nanopore MinION top".
Oxford Nanopore MinION top open
. Source. Before sequencing the DNA, we will do a PCR with primers that fit just before and just after the 16S DNA, in well conserved regions expected to be present in all bacteria.
The PCR replicates only the DNA region between our two selected primers a gazillion times so that only those regions will actually get picked up by the sequencing step in practice.
Eukaryotes also have an analogous ribosome part, the 18S region, but the PCR primers are selected for targets around the 16S region which are only present in prokaryotes.
This way, we amplify only the 16S region of bacteria, excluding other parts of bacterial genome, and excluding eukaryotes entirely.
Despite coding such a fundamental piece of RNA, there is still surprisingly variability in the 16S region across different bacteria, and it is those differences will allow us to identify which bacteria are present in the river.
The variability exists because certain base pairs are not fundamental for the function of the 16S region. This variability happens mostly on RNA loops as opposed to stems, i.e. parts of the RNA that don't base pair with other RNA in the RNA secondary structure as shown at: Code 1. "RNA stem-loop structure".
This is how the 16S RNA secondary structure looks like in its full glory: Figure 5. "16S RNA secondary structure".
Since loops don't base pair, they are less crucial in the determination of the secondary structure of the RNA.
The variability is such that it is possible to identify individual species apart if full sequences are known with certainty.
With the experimental limitations of experiment however, we would only be able to obtain family or genus level breakdowns.
Physics and the illusion of life Updated 2025-12-13
The natural sciences are not just a tool to predict the future.
They are a reminder that the lives that we live daily are mere illusions, religious concepts such as Maya and Samsara come to mind.
We as individuals perceive nothing about the materials that we touch every day really work, nor more importantly how our brain and cell work.
Everything is magic out of our control.
The natural sciences allow us peek, with huge concentrated effort, into tiny little bits a little of those unknowns, and blow our minds as we notice that we don't know anything.
For all practical purposes in life, there is a huge macro micro gap. We are only able to directly perceive and influence the macro events. And through those we try to affect micro events. Because for good or bad, micro events reflect in the macro world.
It is as if we live in a different plane of existence above molecules, and below galaxies. The hierarchy of Figure "xkcd 435: Fields arranged by purity" puts that nicely into perspective, shame it only starts at the economical level, not going up to astronomy.
The great beauty of science is that it allows us to puncture through some of the layers of reality, either up or down, away from our daily experience.
And the great beauty of artificial intelligence research is that it allows to peer deeper into exactly our layer of existence.
Every one or two weeks Ciro Santilli remembers that he and everything he touches are just a bunch of atoms, and that is an amazing feeling. This is Ciro's preferred source of Great doubt. Another concept that comes to mind is when you see it, you'll shit bricks.
Perhaps, the feeling of physics and the illusion of life reaches its peak in molecular biology.
Just look at your fucking hand right now.
Do you have any idea of each of the cells in it work? Isn't is at least 100 times more complex than the materials of the table you hand is currently resting on?
This is the non-science fiction version of the lotus-Eater Machine.
Alan Watts's "Philosopher" talk mentions related ideas:
The origin of a person who is defined as a philosopher, is one who finds that existence itself is exceedingly odd.
The toddler of a friend of Ciro Santilli's wife asked her mum:Our perception of the macroscopic world is so magic that children have to learn the difference between living and non-living things.
James Somers put it very well as well in his article I should have loved biology by James Somers, this quote was brought to Ciro's attention by Bert Hubert's website[ref].The same applies to other natural sciences.
I should have loved biology but I found it to be a lifeless recitation of names: the Golgi apparatus and the Krebs cycle; mitosis, meiosis; DNA, RNA, mRNA, tRNA.In the textbooks, astonishing facts were presented without astonishment. Someone probably told me that every cell in my body has the same DNA. But no one shook me by the shoulders, saying how crazy that was. I needed Lewis Thomas, who wrote in The Medusa and the Snail:For the real amazement, if you wish to be amazed, is this process. You start out as a single cell derived from the coupling of a sperm and an egg; this divides in two, then four, then eight, and so on, and at a certain stage there emerges a single cell which has as all its progeny the human brain. The mere existence of such a cell should be one of the great astonishments of the earth. People ought to be walking around all day, all through their waking hours calling to each other in endless wonderment, talking of nothing except that cell.
Alan Watts' "Philosopher" talk (1973)
Source. Lecture given at UCLA on 1973-02-21. Some key quotes from the talk:The origin of a person who is defined as a philosopher, is one who finds that existence itself is exceedingly odd.
Promoter (genetics) Updated 2025-07-16
Protein tag Updated 2025-07-16